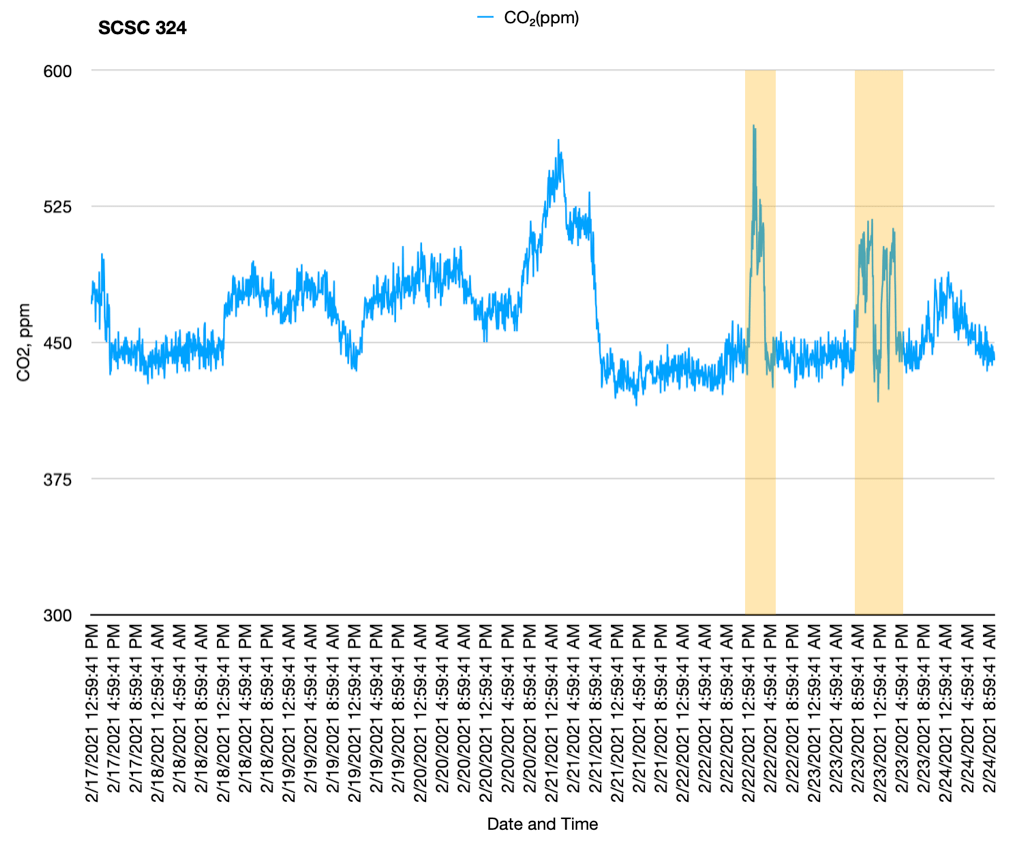
I decided to simplify things by going back to Google Drive. Using OneDrive or Sharepoint or whatever it was turned out to be one tool too many to keep track of. I had made the move from Google Drive to OneDrive a couple years ago when I realized that Google Drive was stamping our time-lapse image sets with the date and time of upload, getting in the way of one of the ways I like to confirm the frequency of image capture for an experiment. It was still possible to get the actual image capture date from the EXIF info, but you couldn’t just sort by that within Google Drive. OneDrive didn’t do that, and we were experimenting with OneNote for lab notes, so it seemed like a good move. Now we’re no longer using OneNote because notebooks were not preserved when students graduated! Fortunately I had backups of notebooks, but this made it untenable.
I feel like using the Google shared drive is better supported by our IT support, and that’s worth something alone. I think the file date problem can be circumvented by just checking the image EXIF info if I need to double-check the interval between images, so even though it isn’t as easy as looking at the file date, at least it’s not lost permanently. Plus students are already using the shared drive to keep their notes and other info, so it’s easier for them to just use the same for data.
The students just log into their student Google account and can access the shared lab drive from there. When I moved things back, I used Transmit, the FTP client from Panic, to log into Google Drive and move large sets of files and folders easily. I think it might be a little more fault-tolerant than the web uploading/downloading interface.